Today I ran FTIR spectroscopy on three ages of DDW, two ages of D2O and the four types of DI that Anthony is using in his current DDW5 experiments.
Many thanks to Stephen Myers for training me on the use of the FTIR, and Dr. Sanjay Krishna for providing me with access to the lab and machine. Stephen was in the lab today and was graciously helpful, as always.
As always I started with a scan of the empty cuvette for the background. Today I kept getting a really strange scan image. Stephen took a look and said confirmed that the scan didn’t look quite right. He changed the Bench Set Gain option to autogain, Anthony cleaned the cuvette, and the next scan came out as we expected it should. After we got a correct background we did a comparison of the previous FTIR background (2/2/2012), today’s background with no autogain correction, and the corrected autogain background. We used today’s autogain corrected background scan for all of our scans today.
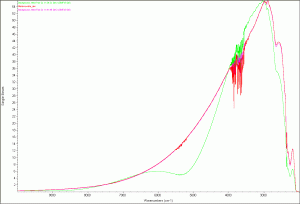
Comparison background scans
For each of the following scans we used the same set up as my previous scans from 2/2/2012, using a quartz cuvette and 3µL of the specified water sample.
My first set of scans were three samples of DDW, each opened on a different date (9/6/2011, 1/17/2012, and 2/16/2012). I expected to see a difference because of possible atmospheric absorption of D2O. What I found was they were all pretty much the same the first scan that I ran. Unfortunately, I messed up the save process and had to re-scan the September and January samples. That scan produced a different result for January as shown in the following figure.
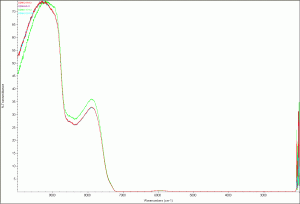
DDW second scan

DDW (second scan) zoomed image
Next I scanned the four types of DI water Anthony is currently using in his current DDW5 experiment (see link). The four types of DI water are:
- DI from the Easypure RoDI (Thermohe) machine in our lab (Ro_DI – purple)
- CHTM’s DI (CHTM_DI – Red)
- Sigma molecular biology grade water (SMol_DI – light blue)
- Sigma double purified water (SDP_DI – green)
The results are surprising. The Sigma double purified water’s scan was slightly different than the other three, which were almost identical. This is certainly something to take a look at.
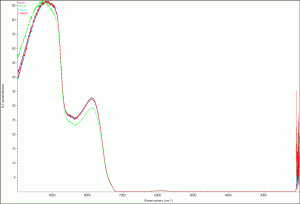
Deionized water scan
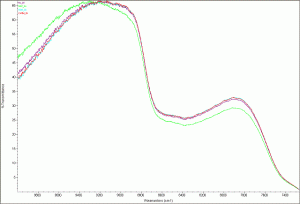
Deionized water - zoomed image
Next I scanned two samples of D2O:
- a bottle opened on 2/16/2012 (red in the scan image)
- a bottle opened on 11/1/2011 (blue in the scan image)
This scan also produced differing results, possibly from atmospheric absorption (what we had expected to see with the different aged DDW).

- D2O scan

D2O scan zoomed
Finally just for comparison, I opened a new window and opened the scan of the February DDW and D2O, the Sigma double processed, and the Sigma molecular biology grade water just to see how the results compared. Interesting that the DDW and Sigma double purified water are identical. Hmmm…

Comparison of DDW, D2O and both of the Sigma DI scans
I’ve uploaded all the raw data onto FigShare with all the images.
My next project is to read up on water frequency and figure out what all the numbers mean. I’ve found a few papers and a website that will probably help shed some light on my very pretty graphs. I’ve ordered two of the papers from the library and the other information is available online.